

PONDEROSA-C/S consists of three programs. User only needs to install
Ponderosa Client for job submission and Ponderosa
Analyzer for validation and refinement. Ponderosa
Server only needs to be installed if you want to build your own
server
on your computational resources (many prerequisites needed with valid
CYANA license). [Click here to move to the download
section!]
Ponderosa Client program is a tool for submitting your input to the
Ponderosa Server. This is a must-have program to run
PONDEROSA-C/S
even though you are thinking of manual refinment and are not planning
to use Ponderosa
Analyzer.
Your contact information
must be filled in correctly to receive the results. You will
receive an e-mail notification that your job submission has been
received by the server a couple of minutes after you click the
“Calculate Strc” button. If this message is not received, it means that
your submission failed to arrive or was denied by the server for some
reason, so please contact the server administrator or Dr.
Woonghee Lee.
Supported chemical shift file
formats
are XEASY prot (.prot),
BMRB NMR-STAR (.str),
SPARKY shift (.shifts),
SPARKY resonance list (.list) and
PIPP (.pipp).
Ponderosa Client automatically generates an XEASY
prot
format file (.prot).
Many sequence file formats
are supported by Ponderosa Client. General 3-letter-code with indices (.seq)
pairs with XEASY prot is accepted. FASTA (.fasta), or
3-letter-code or
1-letter-code without indices in individual lines (.txt)
are also supported. Ponderosa Client automatically generates
3-letter-code with indices format (.seq) from the other formats. For
disulfide bonds and RDCs, Ponderosa Server automatically modifies the
sequence file (CYS to CYSS, pseudo-linkages). Note that if you use
.fasta or .txt, the first sequence index is assigned to 1.
Supported NOESY formats are
two sorts: 1) raw NOESY spectra
(.ucsf, .pipe , .nv or processed Bruker 3rrr),
2) unrefined peak lists (.peaks
or .list).
If
the input is raw NOESY spectra, Ponderosa Client starts by identifying
peaks and conducts an initial peak refinement. This takes seconds to
minutes. Once it is completed, all the peaks saved in the working
directory, and the process is not repeated whenever you change the
settings or other input. This step is necessary because raw NOESY
spectra require much more storage than peak lists. If peak lists are
supplied instead of raw NOESY spectra, this step is skipped, and only
the initial refinement filter will be applied.
Four types of constraints are supported: 1) torsion angle constraints (.aco), 2) distance constraints both upper limits (.upl) and lower limits (.lol), 3) residual dipolar couplings (.rdc), 4) SAXS raw data (.dat) and 5) Ponderosa Blacklist (.blt), Whitelist (.wlt). They are all optional. The Ponderosa Server will automatically generate .aco / .upl / .lol / .tbl files by use of PONDEROSA internal algorithms and/or TALOS / CYANA external modules. The user can provide hydrogen bond restraints in .upl and .lol formats.
Disulfide bonds will be
considered automatically by Ponderosa Server, so please use regular CYS
nomenclature not CYSS in your sequence file.
Weighting factors These are
set to the default value of 50 for the initial run. If you think the
resulting structures are biased, you can change them for subsequent
runs.
Performance options can be
set by the user in Ponderosa Client, but performance options in
Ponderosa
Server have higher priority.
There are eight assignment options:
1) CYANA automation: uses the
CYANA
NOESY assignment module to generate distance restraints and run
structure calculation with them; 2)
PONDEROSA refinement hacks into the CYANA automation and tries
to remove violated constraints and recalculate the structure; 3) Constraints only: does not use
any NOESY spectra or peak lists but only the distance constraints
submitted by the user. This option is used followed by Ponderosa
Analyzer in the final refinement step. 4) AUDANA automation: automated NOE
assignment by AUDANA algorithm in Ponderosa Server and simulated
annealing by Xplor-NIH, 5)
PONDEROSA-X refinement: one additional Xplor-NIH structure
calculation with refined constraints generated by AUDANA algorithm, 6) Constraints only-X: structure
calculation for 40 structures (small scale for fast calculation) with only user uploaded constraints by
XPLOR-NIH, 7) Constraints only-X for
final step with implicit solvation: large number of structure calculation (100 structures) and simulataneous water
refinement by XPLOR-NIH with EEFX potential, 8) Constraints only-X for final step with explicit solvation: large number of structure calculation (100 structures) and following explicit water refinement by XPLOR-NIH.
URL is the location
where the Ponderosa
Server is installed. When you become a NMRFAM user, you will
receive detailed instruction for utilizing our resources.
After submitting the input, you will need to run "Analyze NOESY" for initial peak
picking and refinement. It will take seconds to minutes as mentioned
above. "Calculate strc
" causes the job to be submitted to the Ponderosa Server. After
submission, you no longer need to keep Ponderosa Client open because
all the jobs are carried out on the server side. "Review
results" executes Ponderosa Analyzer.
Screen shot of Ponderosa Client program.
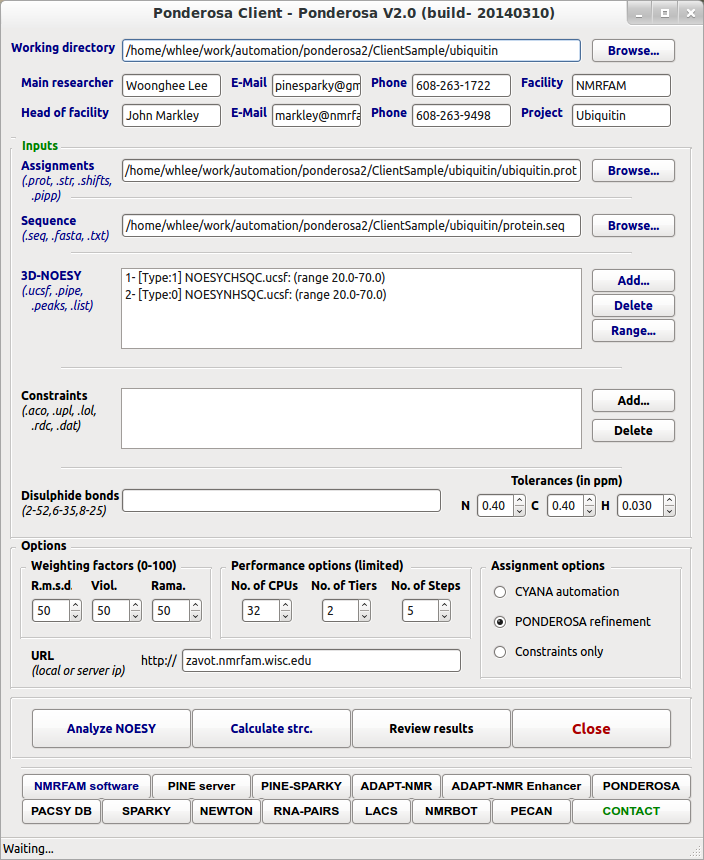
[Jump to the Ponderosa Analyzer]
[Jump to the Download section]
If your institution does not run Ponderosa Server on its site, you do not have to read this section. [Click here to jump to the Ponderosa Analyzer]
To run and maintain Ponderosa Server program on your site, there are some prerequisites to consider.
Operating system must be Linux. NMRFAM runs Ponderosa Server on the CentOS 6 version of Linux.
Have a valid CYANA license, and make it run in any PATH by
setting environmental PATH (optional for CYANA based calculations).
Usually making a symbolic link to the
/usr/local/bin will work. Alternatively (or in parallel), XPLOR-NIH
Ver 2.35 or higher version can be used for Xplor-NIH based
calculations.
STRIDE and any of TALOS versions to be installed on server. They need to be set in environmental PATH, too.
MOLPROBITY enhances the reliability and quality of results. Please install the stand-alone version from here.
XPLOR-NIH
Ver 2.35 or higher, or CNS Ver 1.3
is required to be installed on server with environment PATH setting for
water refinement.
Set up mail. This mailer will send notifications to the users and administrator.
If you have multiple servers to equally distribute jobs, you need to set up NFS (Network File System) to share the files at the same directory names.
Here are the settings of Ponderosa Server program:
Upload directory is where all the data are saved and shared through NFS. Results will be under this directory, too.
External directory is where your apache server allows external access. It will be seen as External URL from outside.
In the External directory contains text files to be sent by the e-mail module to users and administrator. "welcome_email.txt" will be sent when the job is submitted at first, "success_email.txt" and "failure_email.txt" will be sent when the job is succeeded or failed. "admin_email.txt" contains administrator's email address. They will be automatically created when Ponderosa Server is run for the first time.
Screen shot of Ponderosa Server program.
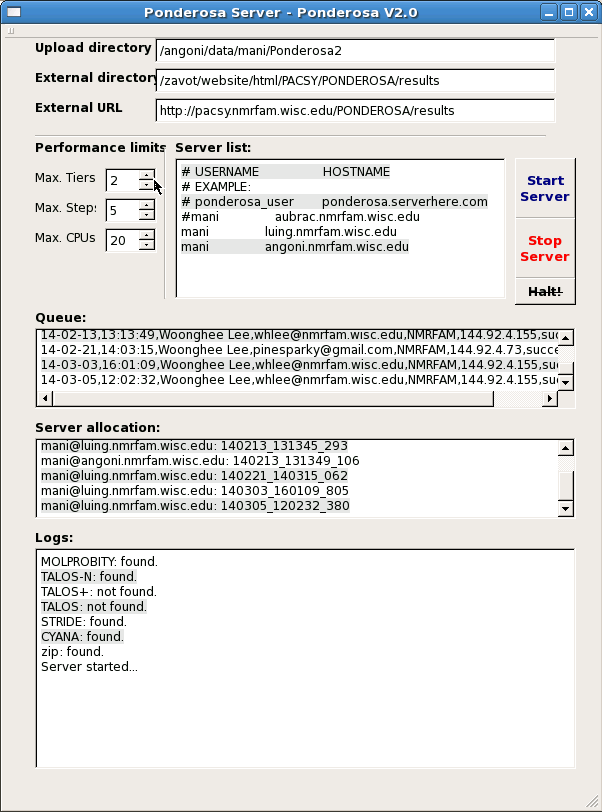
[Jump to the Ponderosa Client]
[Jump to the Download section]
Ponderosa Analyzer is a tool to validate and refine your structure produced by Ponderosa Server. You will receive a compressed zip file after a few minutes to hours after starting the run. PONDEROSA-C/S provides a lot of information regarding your structure, and you may not understand all the directory hierarchies and files. Ponderosa Analyzer is designed to show the important information in the easiest manner for users and provides tools for refinement to achieve a robust, violation-free. publishable structure.
After unzipping the output on your local folder, load the working
directory into Ponderosa Analyzer.
If it is the first time you are working on the output, it will take a
minute for Ponderosa Analyzer to make a complete backup in case you
want to go back to the first output from Ponderosa Server.
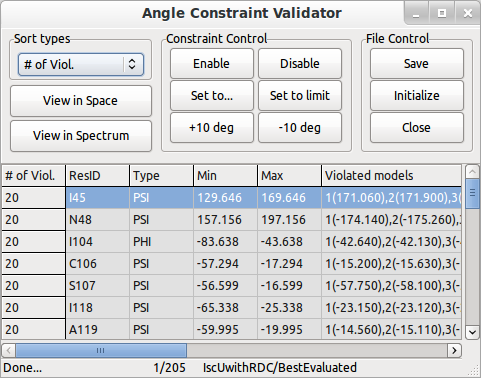
The grid will show calculation directories in each line, and they are categorized by four stages: 1) Best Evaluated (B), 2) Refined (R), 3) Draft (D), 4) Constraints only. Best Evaluatedis the structure with the best score (minimum value) obtained by the Ponderosa Server scoring system. The score includes various factors and so does not necessarily agree with individual factors such as the MolProbity score or target function score. Refined and Draft are about NOESY peak refinement steps during regulating noise threshold for NOESY. You can refer the description of CYCLES and STEPS in the original PONDEROSA document. Constraints only will be present if you used distance constraints as inputs rather than NOESY data. Each line is selectable in the grid, and you can check and correct distance and angle violations by clicking “Show Validation Tools”.
After the correction, you can export your post-processed distance
and angle constraints for a Constraints
only job submission from Ponderosa Client.
Screen shot of Ponderosa Analyzer program.
Screen shot of Distance Constraint Validator from Ponderosa Analyzer.
Screen shot of Angle Constraint Validator from Ponderosa Analyzer.
Ponderosa Violation Investigator
Ponderosa VI (Ponderosa Violation Investigator) is a simplified distance/angle constraint validation program that allows you to investigate violations reported from Ponderosa Server calculation. By integrating the program to PyMOL, user can see selected constraints (multi-selection allowed) in the PyMOL viewer.
We provide the PONDEROSA-C/S
packages. Ponderosa Client and Ponderosa Analyzer in binary
executable formats for Linux (tested on Ubuntu 12.04+ and CentOS 5.5+
with QT 4.8+), Windows and MacOSX (Snow Leopard or newer). Ponderosa
Server is
in binary executable format for Linux only (tested on Ubuntu 12.04+ and
CentOS 5.5+ with QT 4.8+).
* Download
Ponderosa Client [Linux] [Windows] [MacOSX]
Ponderosa Analyzer [Linux] [Windows] [MacOSX]
Ponderosa Server [Linux only]
Ponderosa VI [Linux] [Windows] [MacOSX]
* Installation
Simply, unpack downloaded zip file to the convenient place and enjoy!If it complains about libraries or crashes, just install QT libraries (search for libQtGui) for Ponderosa Client and Ponderosa Server, and Lazarus libraries for Ponderosa Analyzer including GDK-PixBuf libraries (search for libgdk-pixbuf).
* Extended development on this software package
If you want to develop with your novel idea onto our package, contact Prof. John Markley or Dr. Woonghee Lee for details and source codes.
P.S. Old original PONDEROSA is still downloadable from http://ponderosa.sourceforge.net.
However, we highly recommend using the new PONDEROSA-C/S version.
[Jump to the Ponderosa Client]
[Jump to the Ponderosa Server]
[Jump to the Ponderosa Analyzer]